New: The web server to benchmark predictions of 3D genome structure is up an running! You can access the server at https://3dgenbench.net
Aim of the project
Compare the performance of predictive models of chromatin organization and understand which chromatin features define 3D-genome architecture in normal and mutated genomes.
Project overview
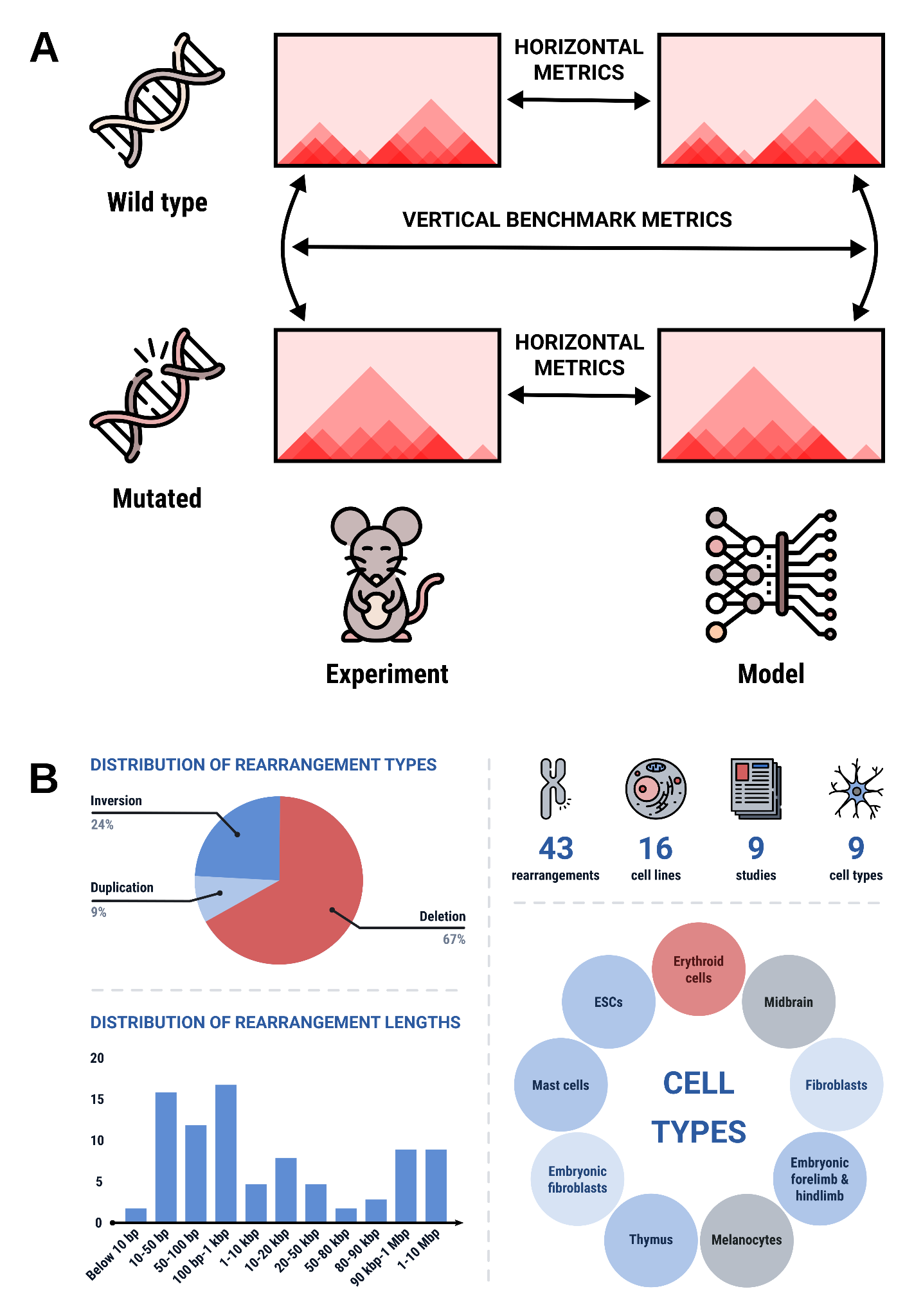
We collected a dataset containing capture Hi-C and complementary ChIP-seq data for wild-type and mutated mouse and human cell types
The current dataset contains samples from several dozen mouse lines harboring genomic mutations with the known effect on chromatin organization, including data from the groups of Stefan Mundlas, Denis Duboule, Douglas Higgs, Laura Lettice, John Rinn and Narimann Battulin. If you have generated 3C-data describing changes of chromatin architecture caused by genetic mutations in human and/or mouse cells and would like us to include your dataset, please get in contact.
The data from all sources is now being re-processed to standardize formats and allow uniform comparisons. The uniformly processed data will be next used by participants to generate models/predict effects of genetic mutations on chromatin architecture. Results submitted by participants will be scored according to the predefined accuracy metrics.
How to contribute
- Join our Slack channel to participate in discussions
- Work with the code at our GitHub repository.
- Test: upload your predictions to the benchmarking server to test your method against experimental data.
- View current experimental datasets
WHAT IS EPIGENETICS?
A video produced by Eskeland laboratory
COST ACTION
COST (European Cooperation in Science and Technology) is a funding organisation for research and innovation networks. Our Actions help connect research initiatives across Europe and beyond and enable researchers and innovators to grow their ideas in any science and technology field by sharing them with their peers. COST Actions are bottom-up networks with a duration of four years that boost research, innovation and careers.
COST
COST (European Cooperation in Science and Technology) is a funding organisation for research and innovation networks. Our Actions help connect research initiatives across Europe and beyond and enable researchers and innovators to grow their ideas in any science and technology field by sharing them with their peers. COST Actions are bottom-up networks with a duration of four years that boost research, innovation and careers.
https://www.cost.eu/
Action Details
- MoU - 108/18
- CSO Approval date - 13/11/2018
- Start date - 13/05/2019
- End date - 13/11/2023
- Former end date - 12/05/2023



© 2024 INC - All rights reserved