INC articles
In this section you will find all the articles that the members of the INC, together, have published. These published works have been supported by the COST Action (CA18127). Please contact us if you are part of our consortium and want to be supported by COST to know all necessary steps you need to follow.
We are also in collaboration with Springer Nature to allow our INC members to publish their work in a special series on [Multiscale Chromatin Dynamics] and co-organized by Clinical Epigenetics (CLEP) and Epigenetic Communications (EPIC). Click on the button below to access all the information needed to participate in this initiative.
2021
Polymer modelling unveils the roles of heterochromatin and nucleolar organizing regions in shaping 3D genome organization in Arabidopsis thaliana
Authors:
DOI: 10.1093/nar/gkaa1275
Abstract:
The 3D genome is characterized by a complex organization made of genomic and epigenomic layers with profound implications on gene regulation and cell function. However, the understanding of the fundamental mechanisms driving the crosstalk between nuclear architecture and (epi)genomic information is still lacking. The plant Arabidopsis thaliana is a powerful model organism to address these questions owing to its compact genome for which we have a rich collection of microscopy, chromosome conformation capture (Hi-C) and ChIP-seq experiments. Using polymer modelling, we investigate the roles of nucleolus formation and epigenomics-driven interactions in shaping the 3D genome of A. thaliana. By validation of several predictions with published data, we demonstrate that self-attracting nucleolar organizing regions and repulsive constitutive heterochromatin are major mechanisms to regulate the organization of chromosomes. Simulations also suggest that interphase chromosomes maintain a partial structural memory of the V-shapes, typical of (sub)metacentric chromosomes in anaphase. Additionally, self-attraction between facultative heterochromatin regions facilitates the formation of Polycomb bodies hosting H3K27me3-enriched gene-clusters. Since nucleolus and heterochromatin are highly-conserved in eukaryotic cells, our findings pave the way for a comprehensive characterization of the generic principles that are likely to shape and regulate the 3D genome in many species.
Differentiating cancer cells reveal early large-scale genome regulation by pericentric domains
Authors:
DOI: 10.1016/j.bpj.2021.01.002
Abstract:
Finding out how cells prepare for fate change during differentiation commitment was our task. To address whether the constitutive pericentromere-associated domains (PADs) may be involved, we used a model system with known transcriptome data, MCF-7 breast cancer cells treated with the ErbB3 ligand heregulin (HRG), which induces differentiation and is used in the therapy of cancer. PAD-repressive heterochromatin (H3K9me3), centromere-associated-protein-specific, and active euchromatin (H3K4me3) antibodies, real-time PCR, acridine orange DNA structural test (AOT), and microscopic image analysis were applied. We found a two-step DNA unfolding after 15-20 and 60 min of HRG treatment, respectively. This behavior was consistent with biphasic activation of the early response genes (c-fos – fosL1/myc) and the timing of two transcriptome avalanches reported in the literature. In control, the average number of PADs negatively correlated with their size by scale-free distribution, and centromere clustering in turn correlated with PAD size, both indicating that PADs may create and modulate a suprachromosomal network by fusing and splitting a constant proportion of the constitutive heterochromatin. By 15 min of HRG treatment, the bursting unraveling of PADs from the nucleolus boundary occurred, coinciding with the first step of H3K4me3 chromatin unfolding, confirmed by AOT. The second step after 60 min of HRG treatment was associated with transcription of long noncoding RNA from PADs and peaking of fosL1/c-myc response. We hypothesize that the bursting of PAD clusters under a critical silencing threshold pushes the first transcription avalanche, whereas the destruction of the PAD network enables genome rewiring needed for differentiation repatterning, mediated by early response bivalent genes.
4D nucleome modeling
Authors: Marco Di Stefano, Jonas Paulsen, Daniel Jost, Marc A Marti-Renom
DOI: 10.1016/j.gde.2020.10.004
Abstract:
The intrinsic dynamic nature of chromosomes is emerging as a fundamental component in regulating DNA transcription, replication, and damage-repair among other nuclear functions. With this increased awareness, reinforced over the last ten years, many new experimental techniques, mainly based on microscopy and chromosome conformation capture, have been introduced to study the genome in space and time. Owing to the increasing complexity of these cutting-edge techniques, computational approaches have become of paramount importance to interpret, contextualize, and complement such experiments with new insights. Hence, it is becoming crucial for experimental biologists to have a clear understanding of the diverse theoretical modeling approaches available and the biological information each of them can provide.
2020
Hi-D: nanoscale mapping of nuclear dynamics in single living cells
Authors: Haitham A. Shaban, Roman Barth, Ludmila Recoules and Kerstin Bystricky
DOI: 10.1186/s13059-020-02002-6
Abstract:
Bulk chromatin motion has not been analyzed at high resolution. We present Hi-D, a method to quantitatively map dynamics of chromatin and abundant nuclear proteins for every pixel simultaneously over the entire nucleus from fluorescence image series. Hi-D combines reconstruction of chromatin motion and classification of local diffusion processes by Bayesian inference. We show that DNA dynamics in the nuclear interior are spatially partitioned into 0.3–3-μm domains in a mosaic-like pattern, uncoupled from chromatin compaction. This pattern was remodeled in response to transcriptional activity. Hi-D can be applied to any dense and bulk structures opening new perspectives towards understanding motion of nuclear molecules.
Coupling chromatin structure and dynamics by live super-resolution imaging
Authors: R. Barth, K. Bystricky, and H. A. Shaban
DOI: 10.1126/sciadv.aaz2196
Abstract:
Chromatin conformation regulates gene expression and thus, constant remodeling of chromatin structure is essential to guarantee proper cell function. To gain insight into the spatiotemporal organization of the genome, we use high-density photoactivated localization microscopy and deep learning to obtain temporally resolved super-resolution images of chromatin in living cells. In combination with high-resolution dense motion reconstruction, we find elongated ~45- to 90-nm-wide chromatin “blobs.” A computational chromatin model suggests that these blobs are dynamically associating chromatin fragments in close physical and genomic proximity and adopt topologically associated domain-like interactions in the time-average limit. Experimentally, we found that chromatin exhibits a spatiotemporal correlation over ~4 μm in space and tens of seconds in time, while chromatin dynamics are correlated over ~6 μm and last 40 s. Notably, chromatin structure and dynamics are closely related, which may constitute a mechanism to grant access to regions with high local chromatin concentration.
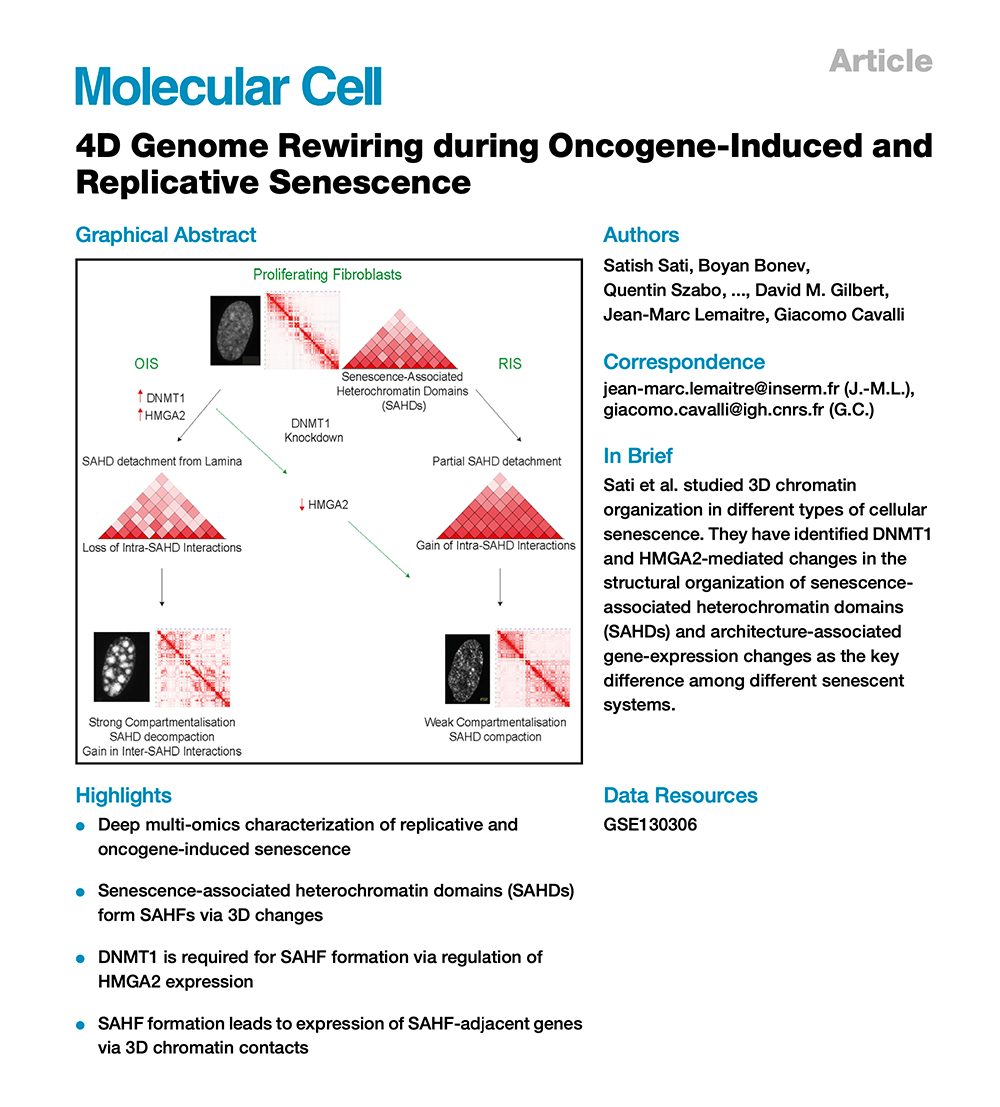
4D Genome Rewiring during Oncogene-Induced and Replicative Senescence
Authors: Satish Sati, Boyan Bonev, Quentin Szabo, Daniel Jost, Paul Bensadoun, Francois Serra, Vincent Loubiere, Giorgio Lucio Papadopoulos, Juan-Carlos Rivera-Mulia, Lauriane Fritsch, Pauline Bouret, David Castillo, Josep Ll. Gelpi, Modesto Orozco, Cedric Vaillant, Franck Pellestor, Frederic Bantignies, Marc A. Marti-Renom, David M. Gilbert, Jean-Marc Lemaitre, and Giacomo Cavalli
DOI: 10.1016/j.molcel.2020.03.007
Abstract:
To understand the role of the extensive senescence-associated 3D genome reorganization, we generated genome-wide chromatin interaction maps, epigenome, replication-timing, whole-genome bisulfite sequencing, and gene expression profiles from cells entering replicative senescence (RS) or upon oncogene-induced senescence (OIS). We identify senescence-associated heterochromatin domains (SAHDs). Differential intra- versus inter-SAHD interactions lead to the formation of senescence-associated heterochromatin foci (SAHFs) in OIS but not in RS. This OIS-specific configuration brings active genes located in genomic regions adjacent to SAHDs in close spatial proximity and favors their expression. We also identify DNMT1 as a factor that induces SAHFs by promoting HMGA2 expression. Upon DNMT1 depletion, OIS cells transition to a 3D genome conformation akin to that of cells in replicative senescence. These data show how multi-omics and imaging can identify critical features of RS and OIS and discover determinants of acute senescence and SAHF formation.
_____
WHAT IS EPIGENETICS?
A video produced by Eskeland laboratory
COST ACTION
COST (European Cooperation in Science and Technology) is a funding organisation for research and innovation networks. Our Actions help connect research initiatives across Europe and beyond and enable researchers and innovators to grow their ideas in any science and technology field by sharing them with their peers. COST Actions are bottom-up networks with a duration of four years that boost research, innovation and careers.
COST
COST (European Cooperation in Science and Technology) is a funding organisation for research and innovation networks. Our Actions help connect research initiatives across Europe and beyond and enable researchers and innovators to grow their ideas in any science and technology field by sharing them with their peers. COST Actions are bottom-up networks with a duration of four years that boost research, innovation and careers.
https://www.cost.eu/
Action Details
- MoU - 108/18
- CSO Approval date - 13/11/2018
- Start date - 13/05/2019
- End date - 13/11/2023
- Former end date - 12/05/2023



© 2024 INC - All rights reserved